Histone Mutations in Cancer

Recent studies show that recurrent somatic mutations in histones (Oncohistones) especially in histone H3 are key drivers in various cancers, including pediatric glioblastoma and soft tissue sarcoma, brain tumors, chondroblastoma, head and neck squamous cell carcinoma, giant cell tumors of bone, and leukemia.
Methods to detect the oncohistones could help in diagnostic and prognostic applications and further the efforts toward targeted therapy. Scientists are already taking advantage of this knowledge to test drugs either targeting the mutation or its consequences.
Overview of five common mutations in histone H3 - 27M, 36M and G34 (R/V/W)
H3K27M is found in 30% of pediatric high-grade glioblastoma (pHGG) and in 80% of diffuse intrinsic pontine glioma (DIPG), and associated with mutations in TP53, PDGFRA, ACVR1, and BCOR. This mutation is also implicated in adult cancers such as glioma, acute myeloid leukemia and melanoma. H3K27M leads to reduced H3K27 methylation, increase in H3K27ac and DNA hypomethylation, and consequently to gene activation with EZH2 inhibition by a negative-dominant effect.
H3K36M is detected in 95% of chondroblastoma, and at lower frequency in pediatric soft tissue carcinoma, head and neck squamous cell carcinoma, melanoma, bladder, and colorectal cancer. H3K36M has a dominant-negative effect on H3K36me2/3, inhibiting the activity of SETD2 and interfering with H3K27 methylation.
H3G34 mutation is found in children and young adult tumors where it is frequently substitute to R or V. H3G34R/V was shown to be correlated with ATRX/DAXX mutation in pHGG. H3G34W and H3G34L mutation is found mostly in the H3F3A gene in 90% of giant cell tumors of bone. H3G34W increases the proliferative and migration capacities of these benign tumors and impairs osteogenic differentiation.
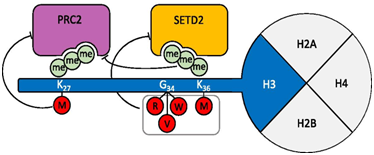
Proposed mechanisms of the main histone H3 mutations. H3K27M leads to a loss of H3K27me3 and H3K27me2 by acting as a dominant-negative inhibitor of PRC2, the complex responsible for H3K27 methylation. H3K36M oncohistone binds and dominantly inhibits the activity of SETD2, the histone methyltransferase responsible for H3K36 methylation. Methylation of H3K36 is known to antagonize the function of PRC2. H3G34 mutants block SETD2 binding, thus reducing its activity on H3K36 methylation. Mutations are indicated in red circles; methyl groups are shown as green circles.
The dark side of histones: genomic organization and role of oncohistones in cancer. Stefano Amatori, et al. Clinical Epigenetics volume 13, 71, 2021

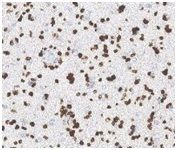
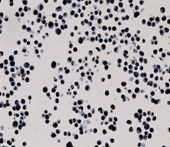
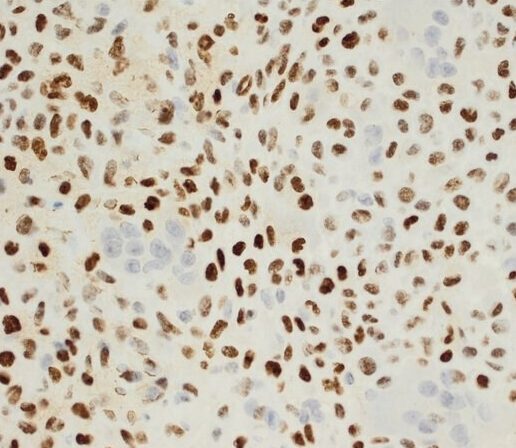

![H3.3G34V [MD258R] RM0223_human giant cell tumor H3.3G34V [MD258R] RM0223_human giant cell tumor](https://medaysis.com/wp-content/uploads/2024/02/H3.3G34V-MD258R-RM0223_human-giant-cell-tumor.png)